Calculate stretch ratio of lattice plane spacings#
Contributor: Martin Diehl (https://martin-diehl.net), Philip Eisenlohr (pe@msu.edu)
DAMASK version: 3.0.2
Prerequisites (data): DADF5 file with deformation gradient (‘F’), elastic deformation gradient (‘F_e’), and crystallographic orientation (‘O’)
[1]:
%matplotlib inline
[2]:
from collections import defaultdict
import damask
import numpy as np
import matplotlib.pyplot as plt
Procedure for lattice stretch calculation#
Since “lattice strains” (actually the stretch of lattice plane spacings) are frequently measured by X-ray diffraction techniques, we adopt the notion that only grains for which a given \(\{hkl\}\) plane is perpendicular (within a tolerance) to a selected reflection direction \(\mathbf{q}\) will contribute to the signal.
For those orientations, the reciprocal stretch ratio \(\lambda_{hkl}^{-1}\) of the \(\{hkl\}\) reciprocal plane spacing can be calculated as
where \(\mathbf{V}_\text{e}\) is the left elastic stretch tensor that acts in the lab frame (following from \(\mathbf{F}_\text{e} = \mathbf{V}_\text{e} \mathbf{R}_\text{cystal}^\text{lab}\)) and \(\mathbf{n}_{hkl}\) is a unit vector along the \(\{hkl\}\) plane normal expressed in the lab coordinate frame.
Explanation#
Let \(\mathbf{d}_{hkl}\) be a vector in the lattice frame that is normal to the lattice plane \(\{hkl\}\) and has magnitude of the reciprocal of the plane spacing. This vector has components \((h,k,l)\) in the dual basis \(\mathbf{e}_j\) of the lattice. Then, \(\mathbf{R}_\text{crystal}^\text{lab}\,\mathbf{d}_{hkl}\), which is parallel to \(\mathbf{q}\), constitutes the same vector but expressed not in the crystal frame but in the lab frame. That plane normal gets transformed by the action of the elastic stretch \(\mathbf{V}_\text{e}\) to \(\mathbf{V}_\text{e}^{-\text T}\,\mathbf{R}_\text{crystal}^\text{lab}\,\mathbf{d}_{hkl} \equiv \mathbf{V}_\text{e}^{-1}\,\mathbf{R}_\text{crystal}^\text{lab}\,\mathbf{d}_{hkl}\). Lastly, the ratio of the reciprocal plane spacings follows as \(\left|\left|\mathbf{V}_\text{e}^{-1}\,\mathbf{R}_\text{crystal}^\text{lab}\,\mathbf{d}_{hkl}\right|\right| \big/ \left|\left|\mathbf{R}_\text{crystal}^\text{lab}\,\mathbf{d}_{hkl}\right|\right|\), which can be simplified to the above expression with \(\mathbf{n}_{hkl} = \mathbf{R}_\text{crystal}^\text{lab}\,\mathbf{d}_{hkl} \big/ \left|\left|\mathbf{R}_\text{crystal}^\text{lab}\,\mathbf{d}_{hkl}\right|\right|\).
Transformation of dual basis#
Let the lattice basis vectors \(\mathbf{e}_j\) be transformed by the linear map \(\mathbf{M}\) such that
This entails the dual basis transforming by the inverse transpose of \(\mathbf{M}\) to
because both transformed bases still maintain biorthogonality, i.e.,
Therefore, the dual basis transforms as
\(\mathbf{e}^{\prime\,i} = \mathbf{R}\,\mathbf{e}^i\) if \(\mathbf{M}\) is a pure rotation \(\mathbf{R} = \mathbf{R}^{-\text{T}}\)
\(\mathbf{e}^{\prime\,i} = \mathbf{V}^{-1}\,\mathbf{e}^i\) if \(\mathbf{M}\) is a pure stretch \(\mathbf{V} = \mathbf{V}^\text T\)
[3]:
def plane_stretch_ratios(F_e,O,hkl,n,tolerance=0,degrees=False):
"""
Calculate stretch ratio of {hkl} planes that are perpendicular to lab direction n from elastic deformation gradient F_e.
Parameters
----------
F_e : numpy.ndarray, shape (...,3,3)
Elastic deformation gradient for each orientation.
O : damask.Orientation, shape (...)
Orientations.
hkl : numpy.ndarray, shape (3)
Miller indices of plane normal.
n : numpy.ndarray, shape (3)
Normal direction in lab frame to select valid orientations having {hkl} aligned with n.
tolerance : float, optional
Maximum deviation angle between lattice plane normal {hkl} and lab normal direction n.
Defaults to 0.
degrees : bool, optional
Tolerance is given in degrees.
Defaults to False.
Returns
-------
ratios : numpy.ndarray (float)
Stretch ratios of all (symmetrically equivalent) {hkl} planes.
"""
hkls = O.to_frame(hkl=hkl,with_symmetry=True,normalize=True)
aligned = np.abs(np.einsum('...i,i',hkls,n/np.linalg.norm(n))) > np.cos(np.radians(tolerance) if degrees else tolerance)
return 1./np.linalg.norm(
np.einsum('...ij,...j',
np.linalg.inv(damask.mechanics.stretch_left(np.broadcast_to(F_e,O.symmetry_operations.shape+F_e.shape)[aligned])),
hkls[aligned],
),
axis=-1)
Selection of diffraction normal and {hkl} planes#
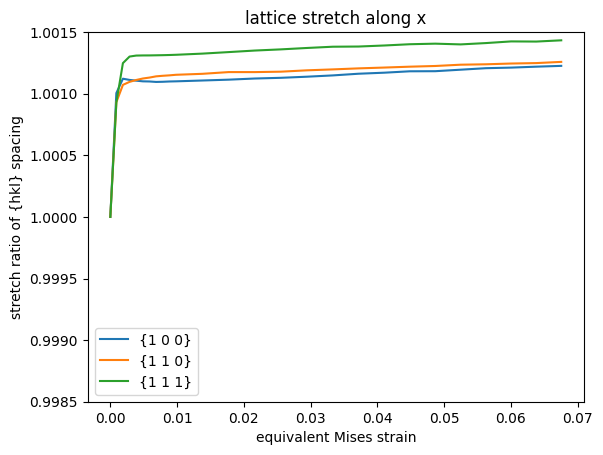
The load case used in this example does unidirectionally extend the sample along \(x\). Hence, interesting directions to interrogate the resulting lattice strains would be along \(x\), \(y\), or \(z\).
The expectation is that lattice plane spacings are generally stretched when measured along \(x\), whereas they should be compressed when observed perpendicular to \(x\), i.e., along \(y\) or \(z\).
Three different lattice planes are selected to examine the influence of directional stiffness and strength of the investigated material, here face-centered cubic Al. For Al, the stiffness is highest along \(\langle 1 1 1 \rangle\) and lowest along \(\langle 1 0 0 \rangle\). Hence, one would expect the lattice plane spacings to be stretched most for \(\{ 1 0 0 \}\) and least for \(\{ 1 1 1 \}\) planes. However, the strength of face-centered cubic lattices is also highest along \(\langle 1 1 1 \rangle\), such that grains oriented as such are prone to attain high stress, which stretches the \(\{1 1 1\}\) spacings relatively more compared to (progressively softer) \(\langle 1 1 0 \rangle\) and \(\langle 1 0 0 \rangle\) orientations than a purely elastic consideration suggests.
[4]:
direction = np.array([1,0,0]) # direction used for lattice plane stretch measurement (Bragg reflection normal) in lab coordinates.
threshold = 5 # allowed misalignment in degree
hkls = [[1,0,0],
[1,1,0],
[1,1,1]] # lattice planes to consider
[5]:
fname = 'lattice_strain/simple_tensionX_material.hdf5'
r = damask.Result(fname)
[6]:
strain = []
stretch = defaultdict(list)
for i in r.increments:
view = r.view(increments=i)
strain.append(damask.mechanics.equivalent_strain_Mises(
damask.mechanics.strain(np.average(view.get('F'),axis=0),t='U',m=0.0)
))
F_e = view.get('F_e')
O_ = view.get('O')
O = damask.Orientation(O_,lattice=O_.dtype.metadata['lattice'])
for hkl in hkls:
stretch[str(hkl)].append(
np.average(
plane_stretch_ratios(
F_e=F_e,
O=O,
hkl=hkl,
n=direction,
tolerance=threshold,
degrees=True)
)
)
[7]:
fig, ax = plt.subplots()
for hkl in hkls:
ax.plot(strain,stretch[str(hkl)],label='{{{} {} {}}}'.format(*hkl))
ax.set_title('lattice stretch along '+'+'.join([f'{a if a!=1 else ""}{d}' for d,a in zip('xyz',direction) if a>0]))
ax.set_xlabel('equivalent Mises strain')
ax.set_ylabel('stretch ratio of {hkl} spacing')
ax.set_ylim(0.9985,1.0015)
ax.legend()
plt.show()